HospitalNetwork-Workflow
Source:vignettes/HospitalNetwork-Workflow.Rmd
HospitalNetwork-Workflow.RmdContext
This R package contains functions to help interested researchers construct networks from data on movement of individual between locations. Although this package was initially developed in the context of networks of healthcare facilities, where the links represent transfer of subjects between facilities, the process can be generalized to the movement of any type of subject between any type of locations.
Formally, a network is composed of nodes which may or may not be connected by edges (or links). In the context of this package, the nodes are the facilities, and the links represent connections between facilities. Although the definition of a node is straightforward, one can define a connection between two facilities in different ways. This package allows to construct a network using various definition of a connection, which are discussed in detail here [ref].
In terms of data structure, a common way of representing a network is with a simple matrix (often called adjacency matrix, or contact matrix). The rows and columns contain the nodes (which appear once in each), and each cell contains the information on whether or not the two nodes are connected.
| A | B | C | D | E | |
|---|---|---|---|---|---|
| A | 0 | 687 | 373 | 296 | 0 |
| B | 0 | 0 | 1294 | 263 | 598 |
| C | 602 | 0 | 0 | 0 | 0 |
| D | 0 | 0 | 718 | 0 | 0 |
| E | 339 | 0 | 86 | 35 | 0 |
At its core, the purpose of this package is to compute the contact matrix from raw data on movement between facilities. Additionally, this package provides the researcher with various tools to analyze and visualize the constructed network.
Data
The package requires a minimal set of information in order to build a network of facilities. This set of data is describe under the section “Required data”. To proceed to further analysis, the package can use additional information in case they are available. These informations are listed under the section “Optional informations”.
Required data
The minimal data needed to construct the network is a simple table with four variables:
- subject ID: an identifier unique to each subject
- facility ID: an identifier unique to each facility
- admission date: the date of admission of the subject in the facility
- discharge date: the date of discharge of the subject from the facility
Therefore, each row must correspond to a unique stay of a subject in a facility. Stays are not allowed to overlap (see Data management).
library(HospitalNetwork)
data = create_fake_subjectDB(n_subjects = 3, n_facilities = 3)
data
#> sID fID Adate Ddate
#> <char> <char> <POSc> <POSc>
#> 1: s1 f3 2019-01-07 2019-01-12
#> 2: s2 f2 2019-01-26 2019-01-28
#> 3: s2 f1 2019-02-12 2019-02-23
#> 4: s2 f1 2019-03-30 2019-04-01
#> 5: s3 f1 2019-01-12 2019-01-15
#> 6: s3 f2 2019-02-13 2019-02-20
#> 7: s3 f2 2019-03-13 2019-03-19Optional data (in the same database)
TODO
| Item | Variable name | Description |
|---|---|---|
| Mode of entry | entry | a variable indicating whether the subject arrived from home (0), or from a facility (1) |
| Mode of discharge | discharge | a variable indicating whether the subject is discharged back home (0), or to a facility (1) |
| Subject residential postcode | postcode | a variable indicating the postal code of the subject residency |
| Wards visited by the subject | ward | a single type of ward predominantly visited by the subject, coded as 1 for ICU or acute care and 0 for others |
Workflow
The main function of the package is
hospinet_from_subject_database(), which takes the database
as argument. It will first performs various diagnostic tests by calling
the function checkBase(), to check for possible issues, and
to ensure that the data is formatted correctly.
If the tests are successful,
hospinet_from_subject_database() will proceed with several
operations and function calls to construct the network. The return value
is an HospiNet R6 object that contains the network itself,
as well as different metrics and information on the network.
Diagnostic tests
The diagnostic tests on the database are performed by the function
checkBase().
The function will check for possible errors in the database. It also
offer the possibility to automatically correct the issues it may have
found in the database. However, since we cannot guarantee having checked
for every possible issue, we encourage you to ensure the data is in the
correct format, and is free of errors, prior to run the function. By
default, checkBase() will not modify the database, but
return informative messages on the issues found. If you wish the
function to try to autocorrect the database, you must set the
corresponding arguments (see Data
management). We recommend that you carefully check the result
afterwards.
# Example
library(HospitalNetwork)
base = create_fake_subjectDB(n_subjects = 100, n_facilities = 10, with_errors = TRUE)
checkBase(base)
#> Error in checkFormat(report = report, convertDates = convertDates, dateFormat = dateFormat, : Dates in Adate are not in Date format.
#> Set argument 'convertDates' to TRUE to convert dates to Date format. Argument 'dateFormat' must be provided as well.Requirements
The requirements to pass the diagnostic tests are the following:
the database must be of class
data.frameordata.table. The functions are implemented in thedata.tableframework, so if adata.frameis provided, it will be converted to adata.table.the database must have at least four columns. By default the column names must be:
c("sID", "fID", "Adate", "Ddate". Although we recommend using these column names, it is possible to use different names by providing them as arguments.columns
"sID"and"fID"must be of typecharacter.dates are handled using functions from the
lubridatepackage. They should bePOSIXct date-time objects. Alternatively, they can be provided as character strings. In that case, you can usecheckBase()to try to parse them to date-time objects usinglubridatefunctions (see Data management).stays should not overlap (see Data management for more details).
Data management
The diagnostic functions of checkBase() will check for
the following issues:
missing values: the following values will be flagged as missing: actual missing values of the form
NAorNaN, character strings"NA", "na", "Na", "N/A", "n/a", "N/a", "NaN"), and empty character strings or empty quotes"", "''".discharge date of a stay anterior to its admission date.
overlapping stays: TODO
You can use the function checkBase() to try to correct
automatically the issues it has found by setting the corresponding
arguments.
missing values: to remove entries with missing values, set the argument
deleteMissing. If set to"record", the record with the missing value will be removed. If set to"subject", all records of the same subject will be removed.errors: to remove errors, such as discharge date of a stay anterior to admission date, set the argument
deleteErrors. If set to"record", the record with the error will be removed. If set to"subject", all records of the same subject will be removed.overlapping stays:
checkBase()will automatically handle overlapping stays as follows: TODOdates: if admission and discharge dates are provided as character strings, you can use
checkBase()to try to parse them to a date-time object, which uses internally thelubridatefunctions. To do that, set the argumentconvertDates = TRUE. You must also specify in what format are the dates (“year-month-day”, “day-month-year”, etc.) by setting the argumentdateFormat = c("ymd", "ydm", "dmy", "dym", "mdy", "myd").
The HospiNet object
HospiNet is an R6 object containing the facility matrix as well as specific information regarding the network. We have developed a summary and a print method for this object. The information contained in an HospiNet objects are:
- n_facilities, the number of facilities in the network,
- n_subjects, the number of subjects in the network,
- n_movements, the number direct or indirect (depending on the window size) transfers,
- window_threshold, the size of the movement window in days (0 for direct transfers between facilities),
- hist_degrees, _in, _out, are named vectors containing the number of nodes for each degree, indegree, or outdegree,
- …
Using the package
mydbmed = create_fake_subjectDB(n_subjects = 100, n_facilities = 10)
hn = hospinet_from_subject_database(base = mydbmed, noloops = FALSE)
#> Input database was not checked yet, which is required for network reconstruction.
#> Running 'checkBase()' with default parameters.
#> If this doesn't work, please run checkBase() separatelty with custom parameters first.
#> Warning in get_hubs_bycluster(graphs = graph_byclust, name =
#> "cluster_fast_greedy"): Cluster(s) clust_4 have only one member
hn
#> 10 facilities and 144 movements.
#> Movement window is 365 days.
#> Constructing full matrix
#> f01 f02 f03 f04 f05 f06 f07 f08 f09 f10
#> f01 4 1 0 3 1 0 0 2 1 0
#> f02 2 1 2 5 2 2 2 0 0 3
#> f03 1 2 1 0 2 1 2 1 3 2
#> f04 1 2 2 0 1 1 2 2 4 2
#> f05 2 0 2 0 1 1 1 1 0 1
#> f06 2 2 1 2 1 1 1 1 0 3
#> f07 1 2 0 1 1 2 1 1 1 2
#> f08 0 1 0 2 1 1 2 2 1 2
#> f09 1 1 2 1 3 2 2 1 3 2
#> f10 2 0 1 3 1 3 0 2 0 4
plot(hn)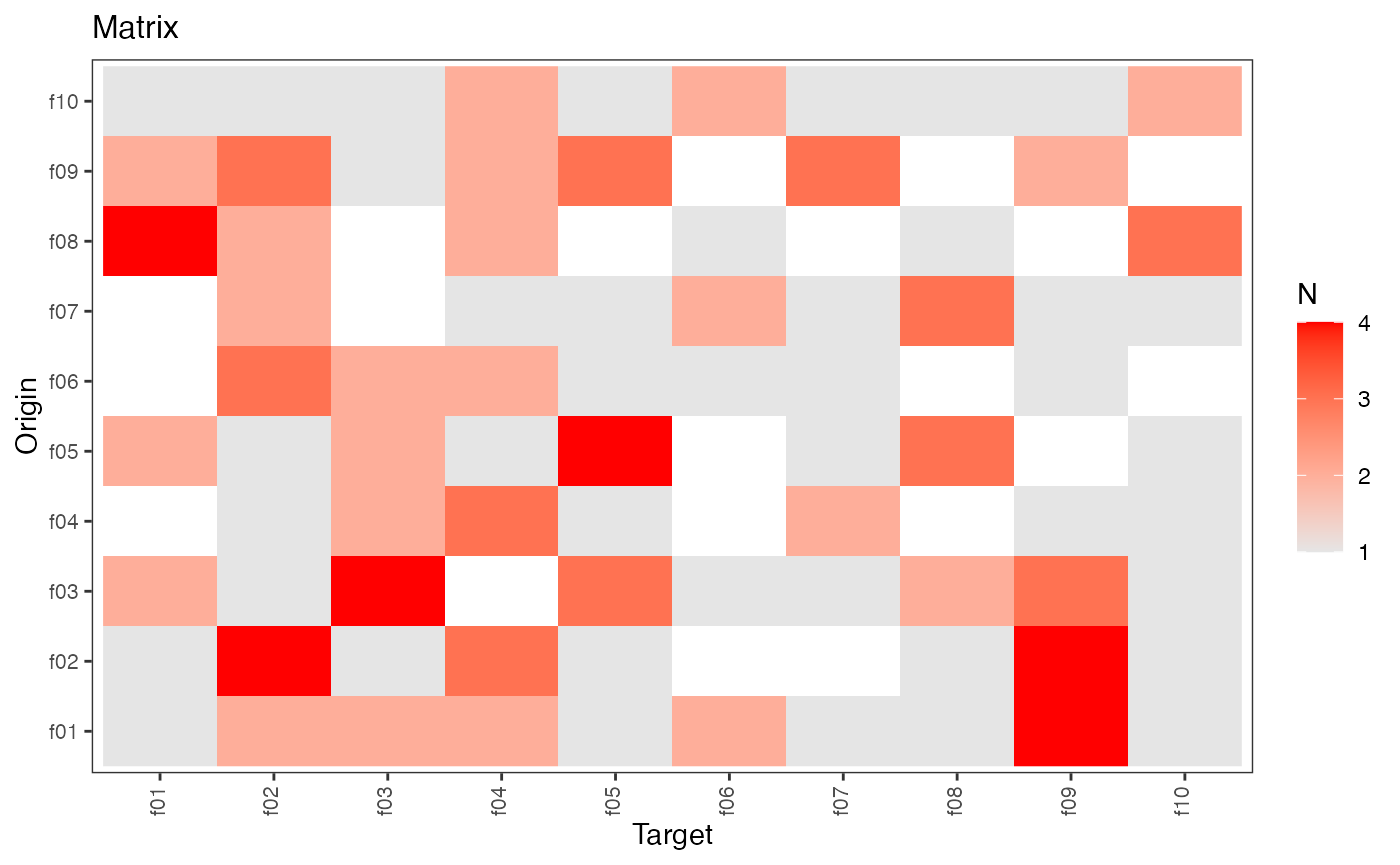
plot(hn, type = "degree")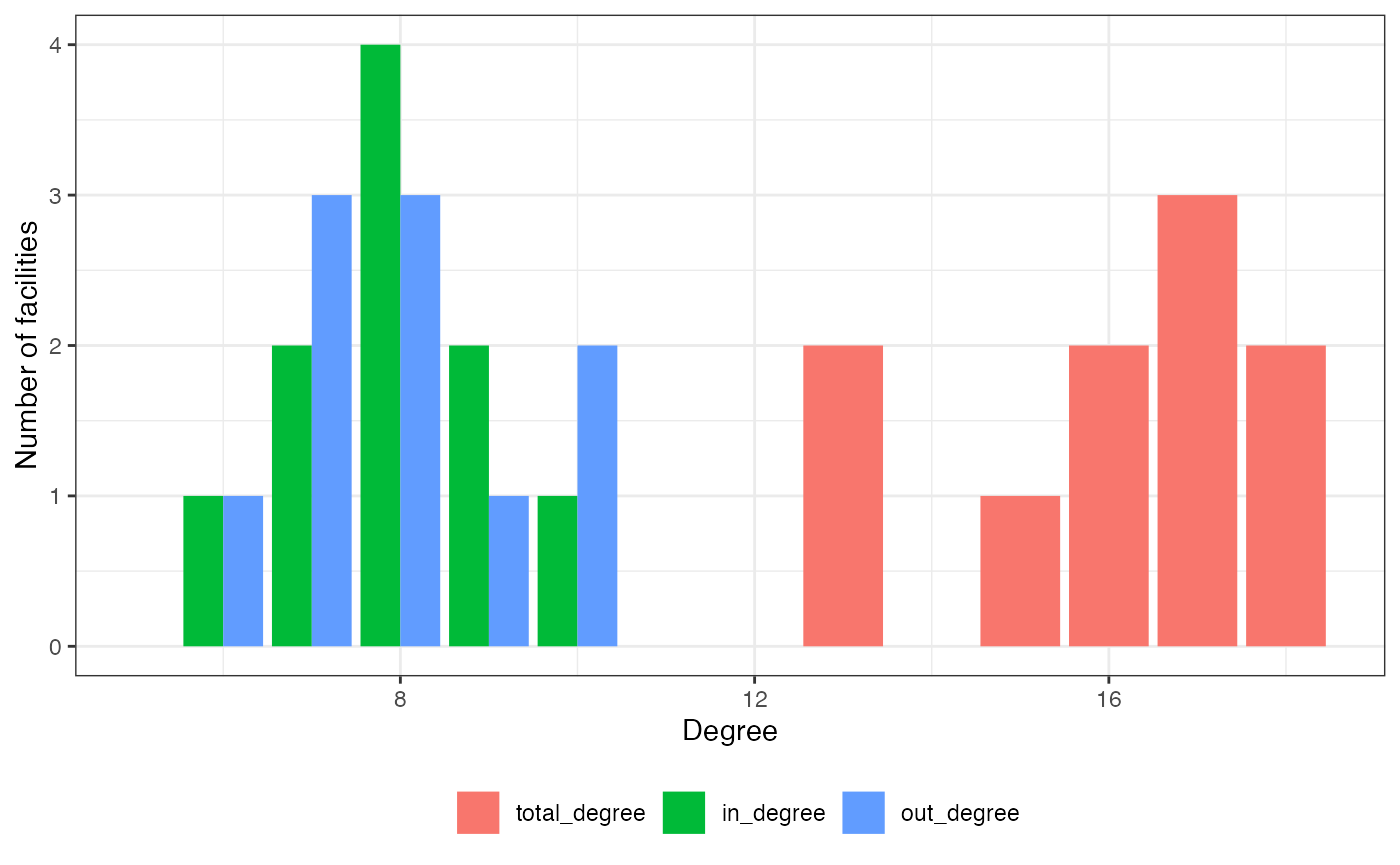
plot(hn , type = "clustered_matrix")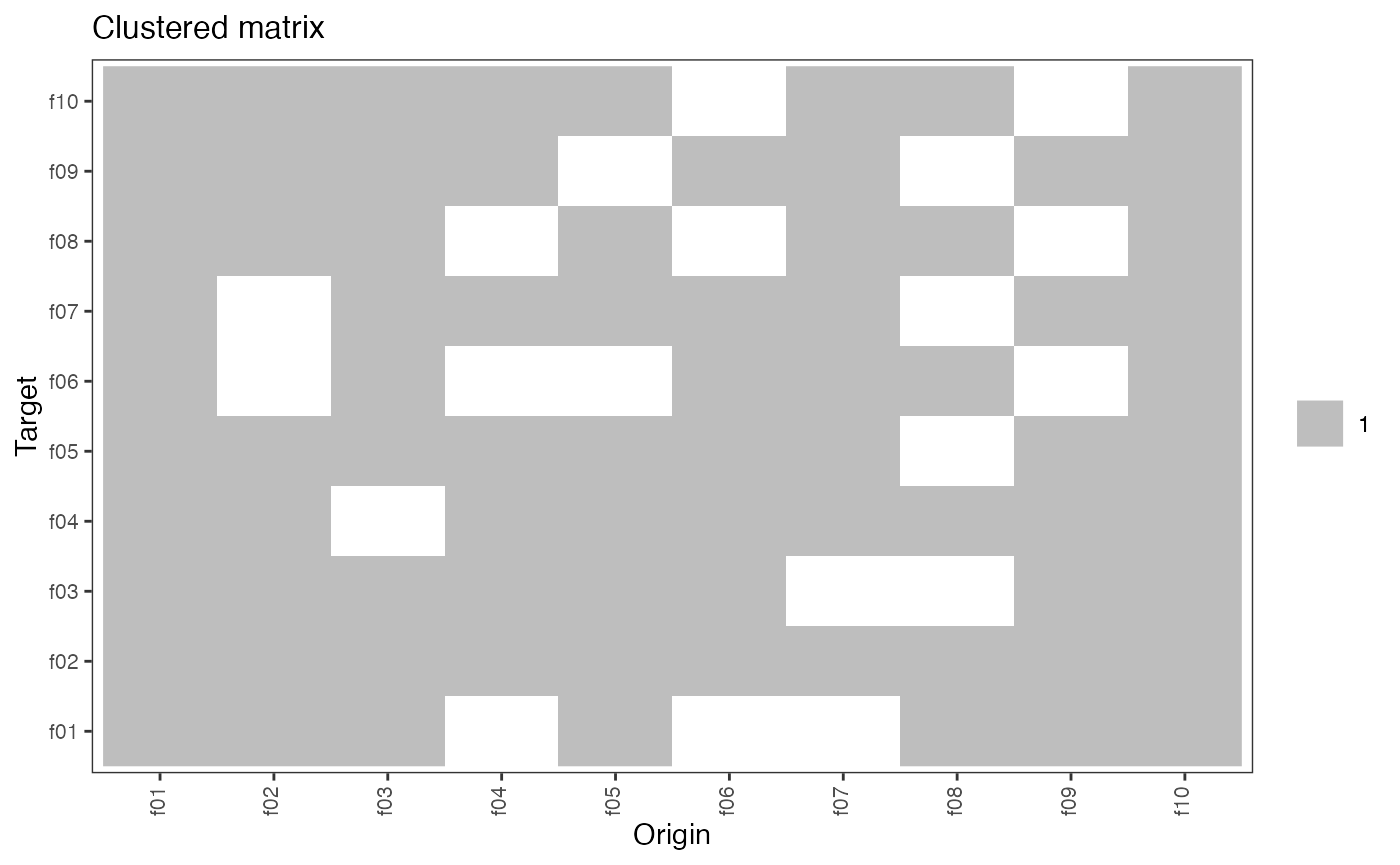
mydb = create_fake_subjectDB_clustered(n_subjects = 10000, n_facilities = 100, n_clusters = 5)
hn = hospinet_from_subject_database(base = mydb, noloops = FALSE)
#> Input database was not checked yet, which is required for network reconstruction.
#> Running 'checkBase()' with default parameters.
#> If this doesn't work, please run checkBase() separatelty with custom parameters first.
hn
#> 100 facilities and 18169 movements.
#> Movement window is 365 days.
#> Matrix too big to be printed on screen.
plot(hn)
plot(hn, type = "degree")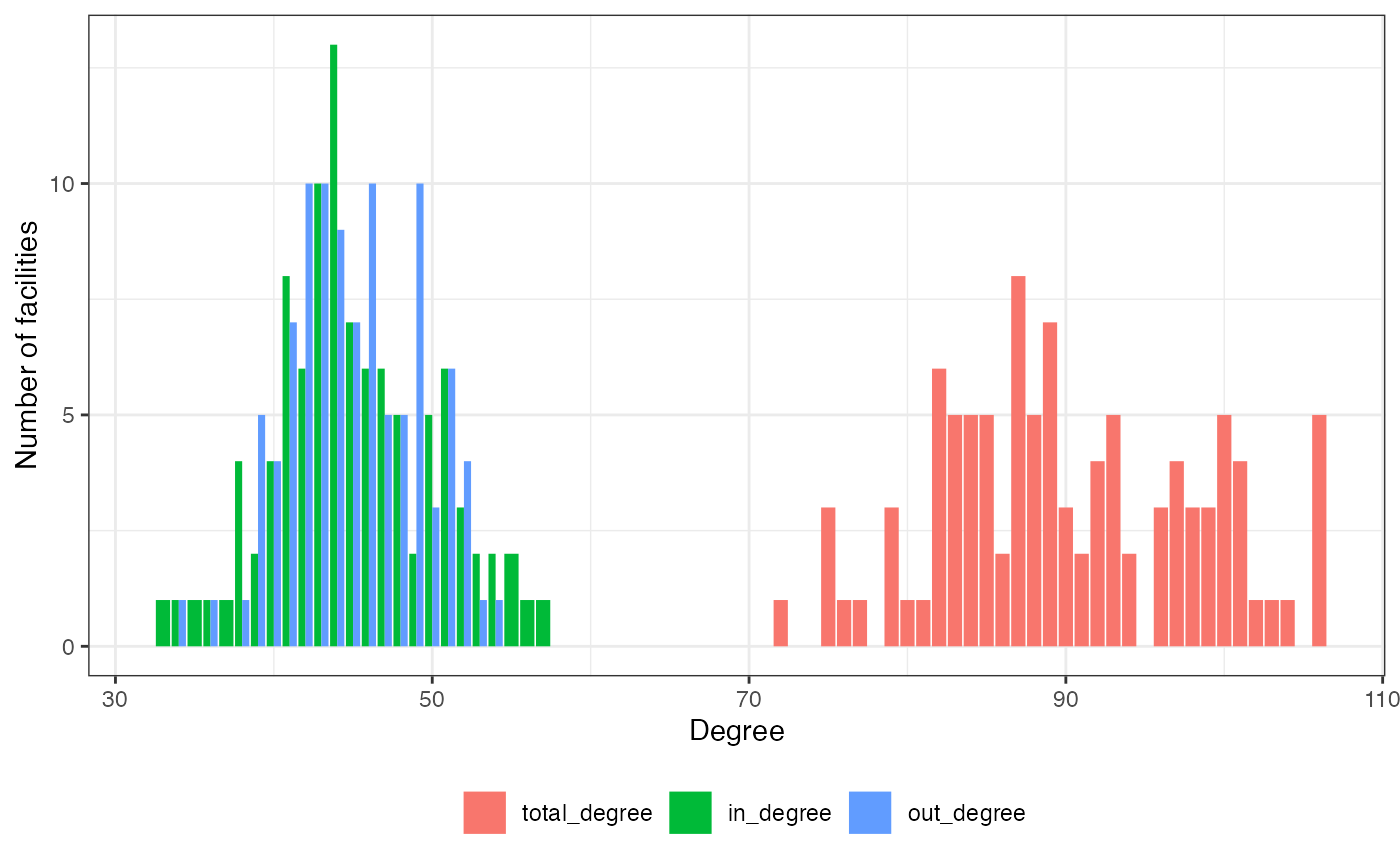
plot(hn , type = "clustered_matrix")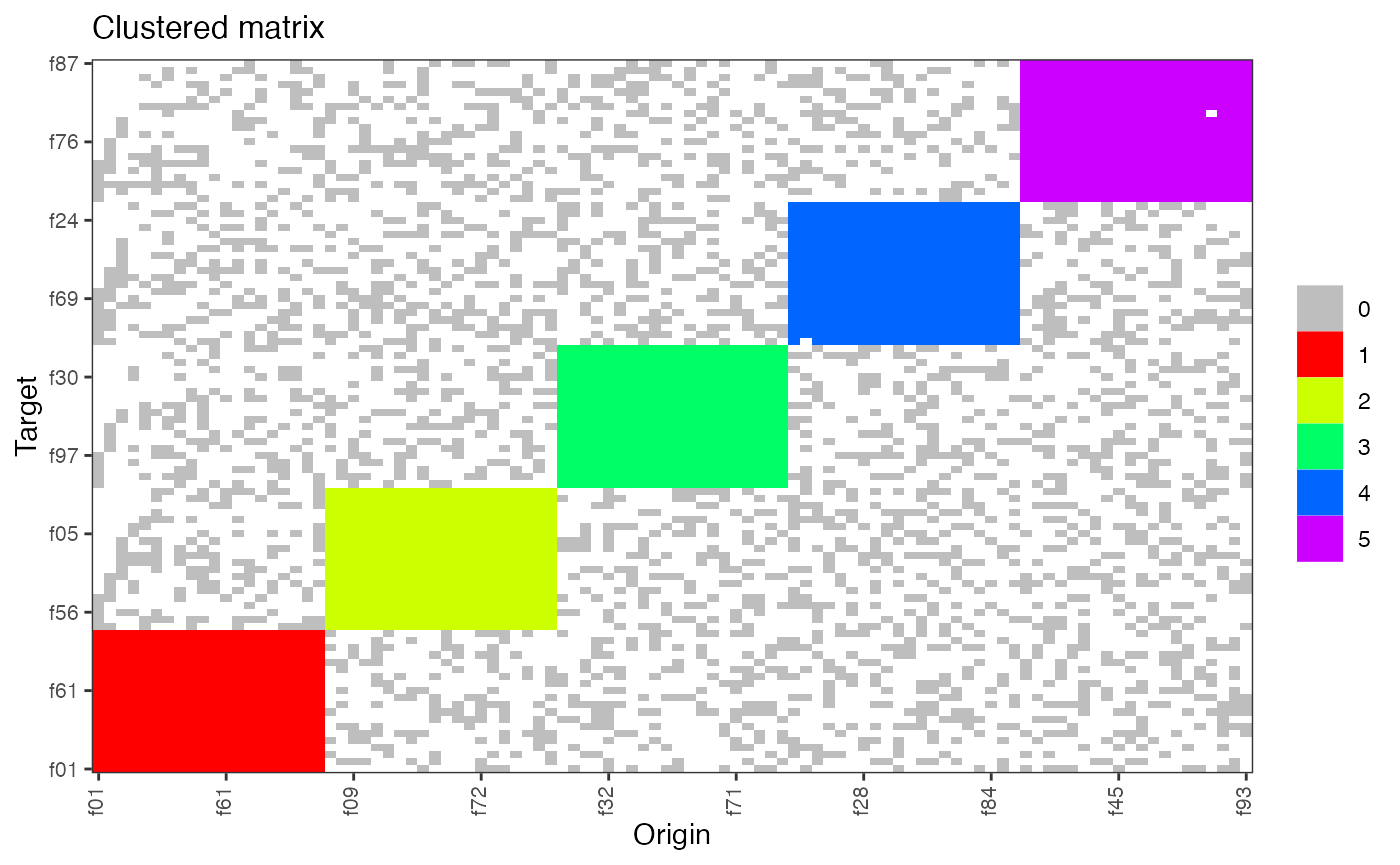
#this plot may not work on some systems
#plot(hn , type = "circular_network")